What is CriSNPr?
CRISPR Diagnostics (CRISPRDx) for CRISPR-based SNV detection refers to a broad category of biochemical for the rapid and effortless detection of a target SNV in a polynucleotide sequence. These assays are majorly designed around the ability of Cas effectors to distinguish nucleic acid sequences on the basis of mismatches at defined positions within the crRNA. Several Cas effectors such as LbCas12a, Cas14a, AaCas12b, and en/FnCas9 exhibit nucleotide position-specific mismatch sensitivity and have previously been employed for SNV detection (1-7). CriSNPr is a web server that helps a user to design position-specific mismatch sensitive modified crRNAs for en/FnCas9, LwCas13a, AaCas12a, LbCas12b, and Cas14a. Additionally, it provides specific primer sequences which when used together with the gRNAs for every Cas diagnostic pipeline, can be advanced for developing rapid CRISPR-based SNV diagnostic assays. CriSNPr begins by mapping the provided SNV information either as an ID or a sequence to the selected genome. Once mapped, a custom scripted algorithm searches for the required protospacer adjacent motif (PAM) near the SNV fitting with criteria for any of the previously reported crRNA mismatch sensitive positions as mentioned in table 1. Using CriSNPr, the modified crRNAs are also checked and reported for any off-targets within the target genome. In addition, the amplification primers are also pre-filtered for off-targets within the human, bacterial and viral genomes available to date at the NCBI genome database.
Table 1, Cas effectors with their mismatch sensitive crRNA nucleotide positions.
| Cas effector | PAM/PFS | Spacer Length (bp) | Cleavage target | Mismatch sensitive positions (From PAM) | Reference |
|---|---|---|---|---|---|
| LwCas13a | NA* | 22-28 | RNA | 3&4, 3&5 | (Cameron et al., 2018) |
| LbCas12a | TTTN | 16-18 | ssDNA/dsDNA | 1st -7td | (Li et al., 2018) |
| Cas14a | NA | 20 | ssDNA/dsDNA* | 11&12, 12 | (Harrington et al., 2018) |
| AaCas12b | TTN | 20 | ssDNA/dsDNA | 1&4, 1&5, 5&8, 4&16, 4&11, 5&11, 16&19, * | (Teng et al., 2019) |
| Fn/enFnCas9 | NGG/NRG or NGR | 20 | dsDNA | 2&6, 16&19, 17, 18, 19 | (Azhar et al., 2021; Kumar et al., 2021, Acharya et al., 2021) |
For clinical relevance, the current version of CriSNPr provides a readymade pipeline for CRISPR-based detection of pathogenic and non-pathogenic mutations for all reported human nucleotide variants (dbSNP) and SARS CoV-2 variants of interest/concern (VOI/VOC). Nevertheless, the crRNA designs and the detection assays can also be developed de novo variants of choice with or without any clinical relevance present within human or SARS-CoV-2 genomes. The current version of CriSNPr contains reference SNVs information from human dbSNP and SARS-CoV-2 CNCB-NGDC (China National Center for Bioinformation-The National Genomics Data Center), respective databases.
Directions for Input and output
CriSNPr, online web-server queries for the SNV of interest and provides users with information regarding all Cas systems that can be used to detect that particular SNV, and the required modified crRNA and primer design parameters based on gRNA design principles available in the literature for each Cas protein
- Human, with ready-made targetable clinically relevant SNPs from the human dbSNP database
- SARS-CoV-2, with ready-made targetable SNVs within the SARS-CoV-2 genome
- Seq-CriSNPr, to be used for sequence-based search of any reported targetable SNV with/without clinical any clinical relevance within the human or SARS-CoV-2 genomes
The current interface of CriSNPr has been divided into three subdomains featuring,

Fig. 1, CriSNPr’s first user page with choice for human, SARS-CoV-2, and Seq-CriSNPr SNV targetable algorithms.
i) HUMAN; for targeting human SNPs/SNVs with pathological relevance
As the current version of CriSNPr contains reference SNVs information from the human dbSNP database, a user can query for designed sequences using the dbSNP rsID for humans.
Fig. 2, Human SNV rsID input window with examples for user’s guide
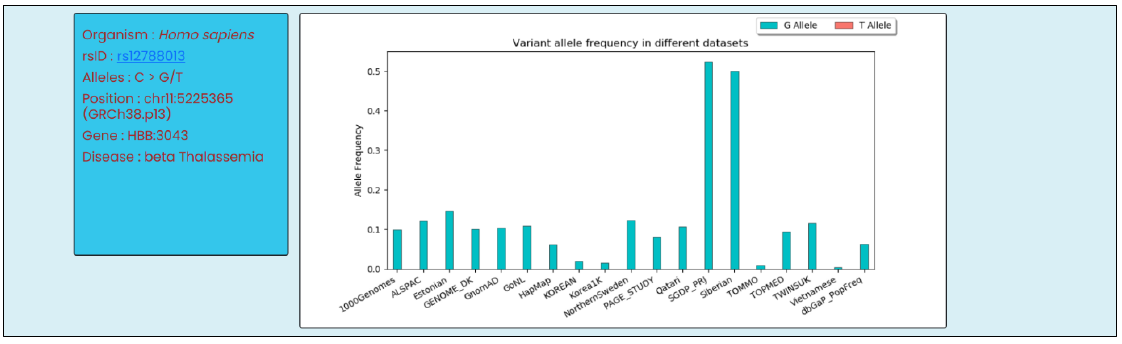
After a user’s entry, the CriSNPr web-server first tries to authenticate the entered input as a valid input by mapping and visualizing SNVs/SNPs, for related information like an organism of origin, genome coordinates, gene ID, disease association, reported allele, and their population-specific frequency distribution. The targetability of a queried SNP/SNV by all included Cas systems is simultaneously checked and only the Cas systems with positive results are shown with the required information.
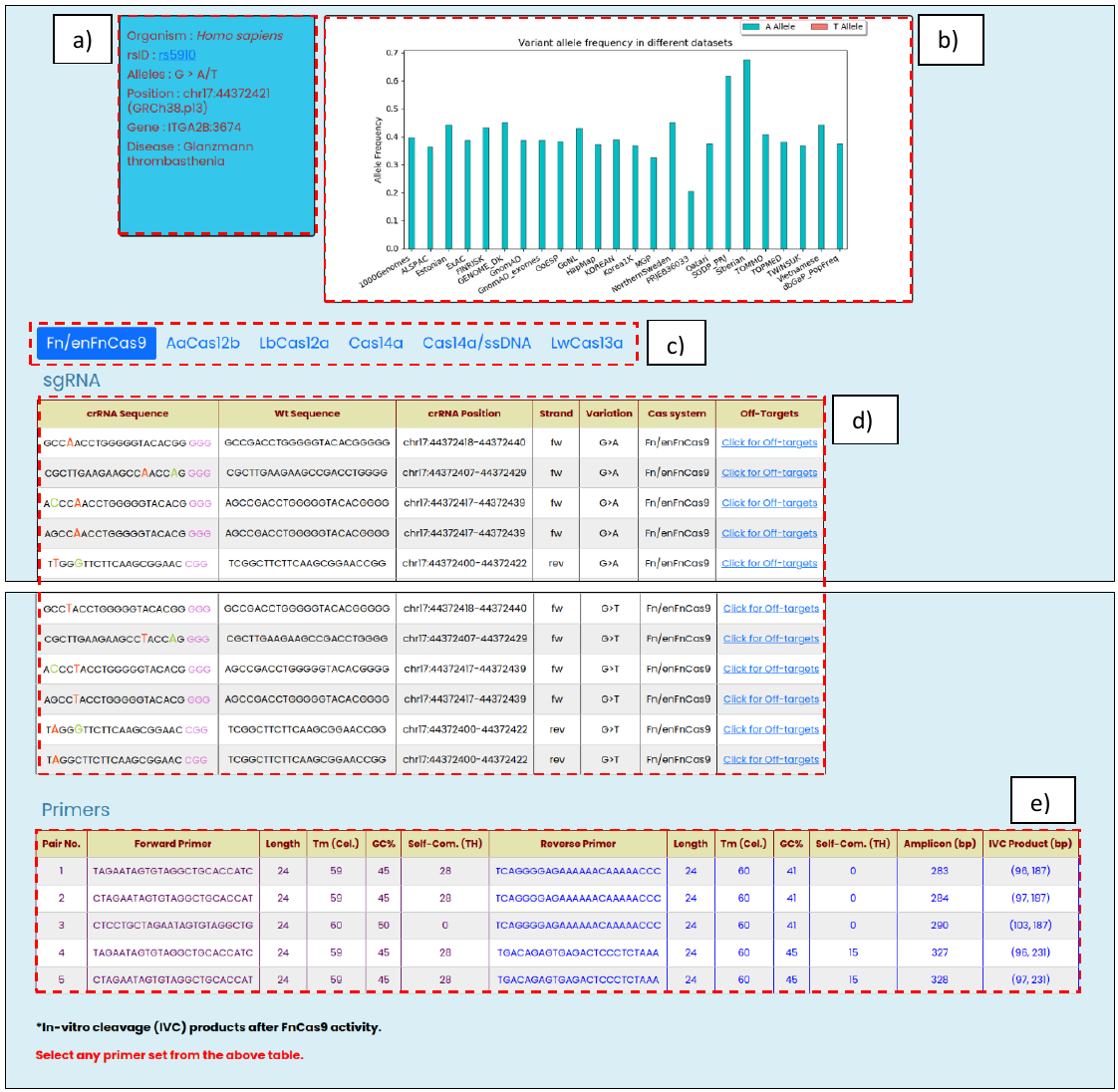
- SNV validation, with information such as an organism of origin, user-provided rsID, reported alleles, genome coordinates, gene ID, disease association
- Population-specific frequency distribution of different alleles reported at that genomic position
- Cas system selection panel highlights the selected option, and results are shown for it
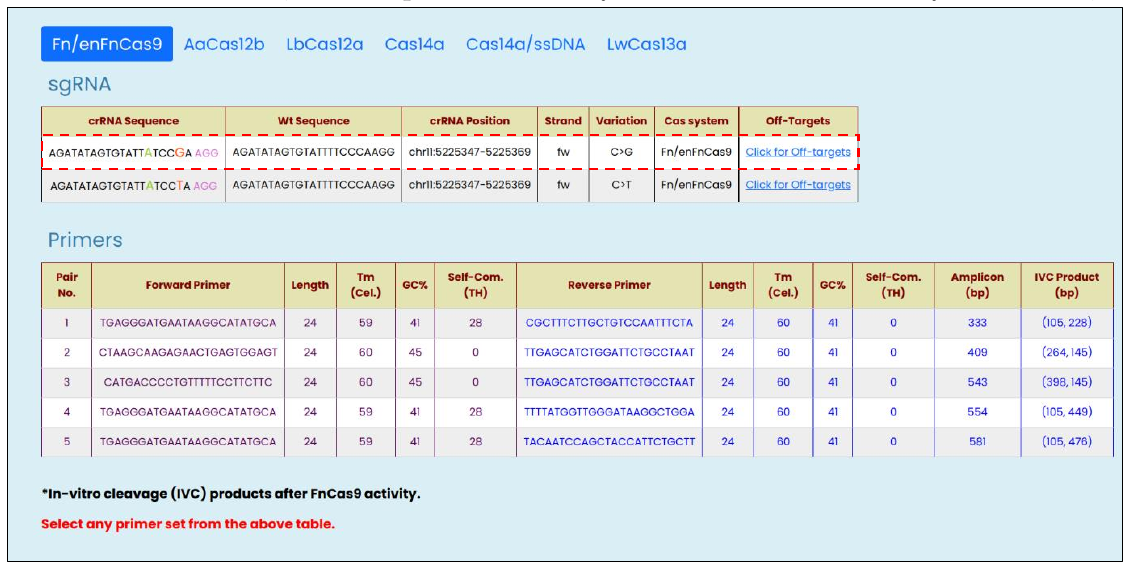
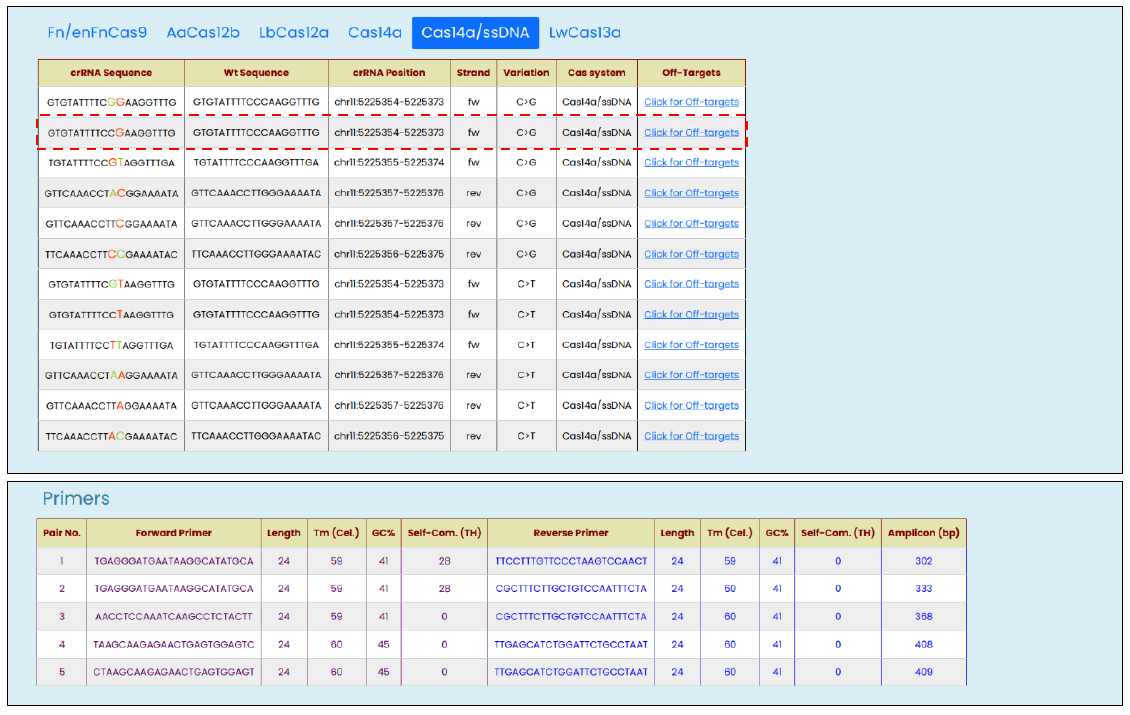
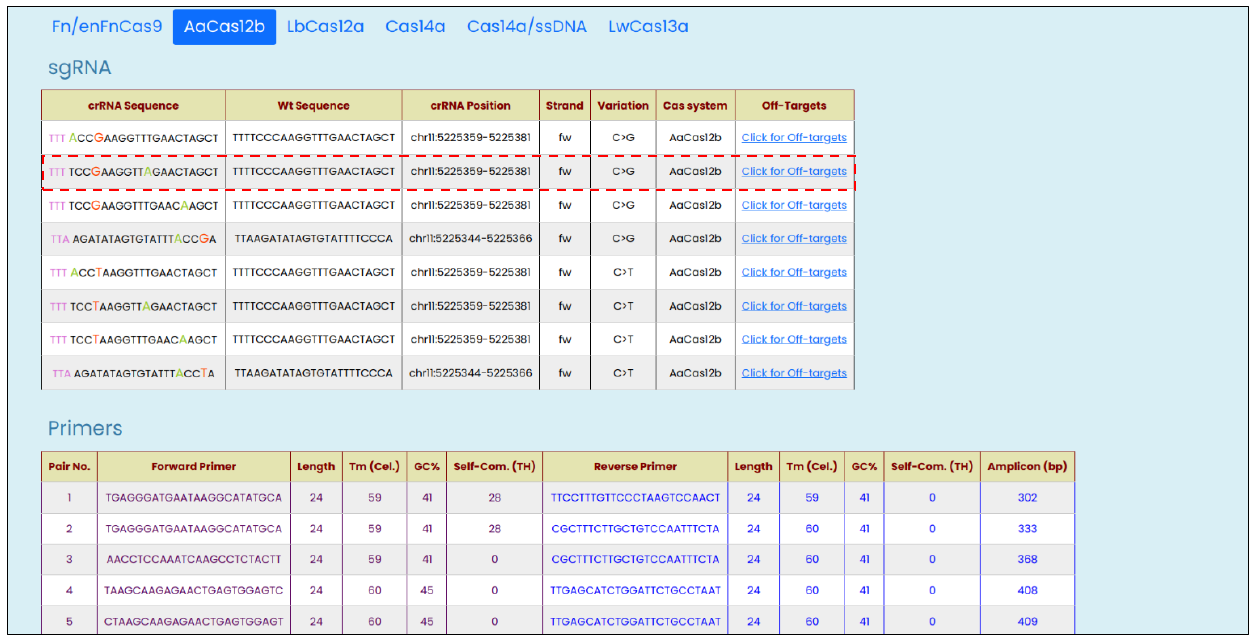
- SNV targeting modified crRNA sequences with other related information such as reference WT sequences, genomic positions, strand orientation, targeted allelic change, and the list of off-targets against the modified crRNA.
- Top 5 PCR amplicon primer-pair with annealing temperature, GC%, and self-complementary (TH) score as provided by the Primer3Plus (8)
Fig. 3, Targetable Human SNV rsID output window with the following information:
Since the position of mismatches differs between different Cas proteins used for designing the diagnostic pipeline, CriSNPr provides also returns information highlighting the position of mutant nucleotide (red color) and synthetic nucleotide (needed depending on the Cas systems used, green color) within a crRNA sequence.
CriSNPr also gives information about the off-targets for the SNV targeting modified crRNA sequences. To identify potential off-targets against the modified crRNAs, CriSNPr takes the advantage of the previously reported offline versatile algorithm of Cas-OFFinder (9). Using this CriSNPr reports off-target information against a modified crRNA as a list containing chromosome location and coordinates of the off-targets, DNA strand orientation, and the number of off-target sequences with up to 4 nucleotide mismatches, etc.
All the primer pairs provided by CriSNPr are also pre-filtered for off-targets with up to 2 nucleotide mismatches against representative bacterial genome database (NCBI), virus genome database (NCBI) (Brister et al., 2015), and human genome/transcriptome (GENCODE GRCh38) (Frankish et al., 2019). Considering SNV detection with clinical relevance or diagnosis, only the primer sequences with very high specificity to minimize possibilities of non-specific amplicons are provided by the CriSNPr. In addition, as FnCas9/enFnCas9 can generate a gel-based readout upon target cleavage, the amplicon primers are specially designed to give cleaved DNA products with a size difference of almost two times than other, to discriminate between wild-type and mutant sequences on an agarose gel.
Subsequently, if any SNV is targetable by other Cas effectors too, CriSNPr reports the designed crRNA and primer sequences for that Cas effector too, once selected by the user.
ii) SARS-CoV-2; for targeting reported SARS-CoV-2 variants
CriSNPr also contains SNV information from the SARS-CoV-2 CNCB-NGDC (China National Center for Bioinformation-The National Genomics Data Center) database. With no prior sequence information about an SNV, a user can access CRISPRdx design-based mutant amino acid position for SARS-CoV-2.
The SARS-CoV-2 interface considers standard variation nomenclature requiring a gene name followed by a “_” (underscore), reference amino acid, position at protein, and alternative amino acid. For example, S_N501Y, S_E484K, etc. Upon finding a match in the CriSNPr database of targetable SNVs, it again gives out SNV identity information along with variation frequencies, crRNAs, and primer oligos for SNV detection in the variant SARS-CoV-2 lineages by the Cas systems integrated into the CriSNPr.
Fig. 4, SARS-CoV-2 mutation input window with examples for user’s guide
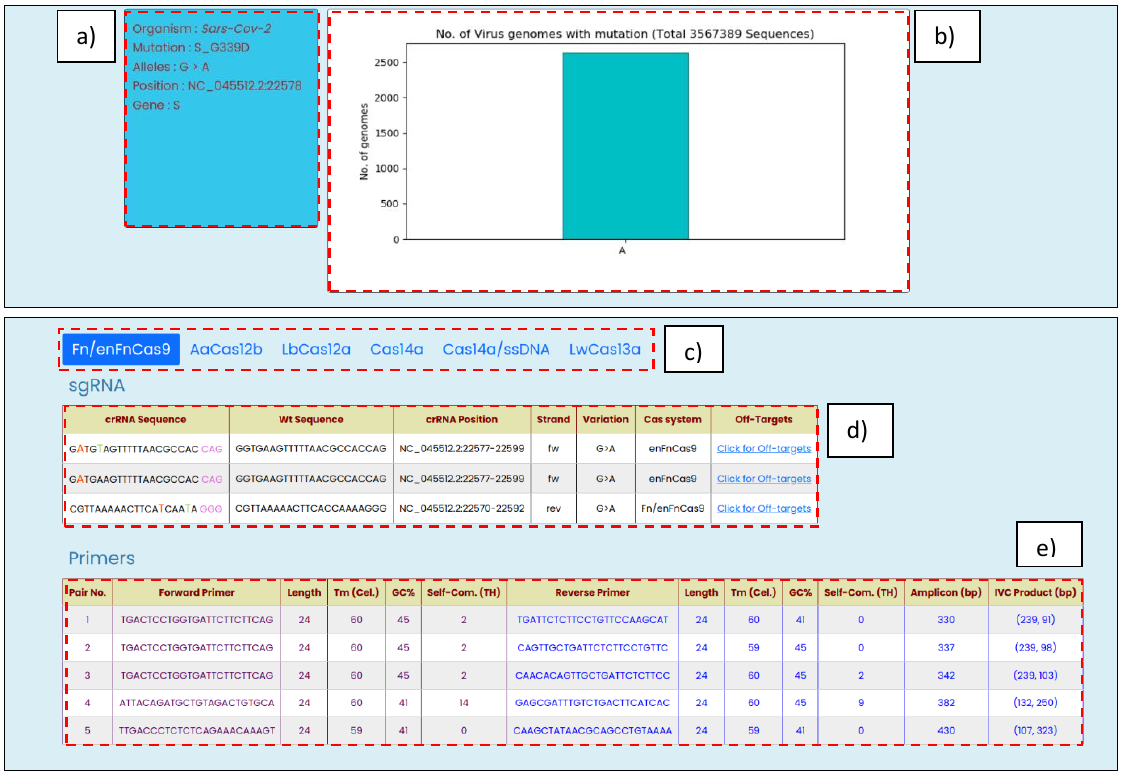
- Mutation validation, with information such as an organism of origin, user-provided mutation, reported alleles, genome coordinates, and gene name.
- Genomic-level frequency distribution of mutation containing SARS-CoV-2 genomes deposited at GISAID database.
- Cas system selection panel highlights the selected option, and results are shown for it
- Mutation targeting modified crRNA sequences with other related information such as reference WT sequences, genomic positions, strand orientation, variation change, and the list of off-targets against the modified crRNA.
- Top 5 PCR amplicon primer-pair with annealing temperature, GC%, and self-complementary (TH) score as provided by the Primer3Plus
Fig. 5, Targetable SARS-CoV-2 mutation output window with the following information:
Like the human interface, the SARS-CoV-2 interface also provides all the necessary information required for developing CRISPR-based variant detection assays.
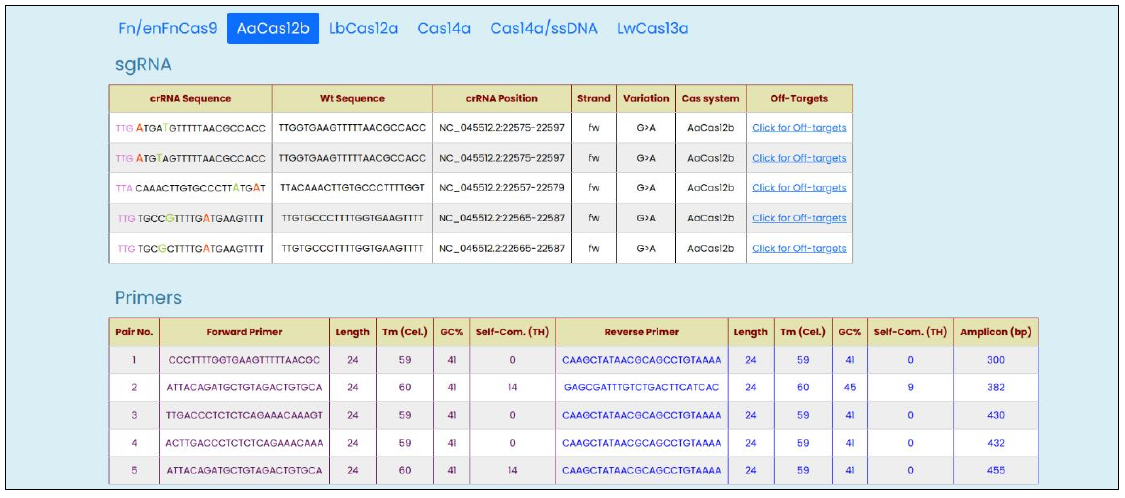
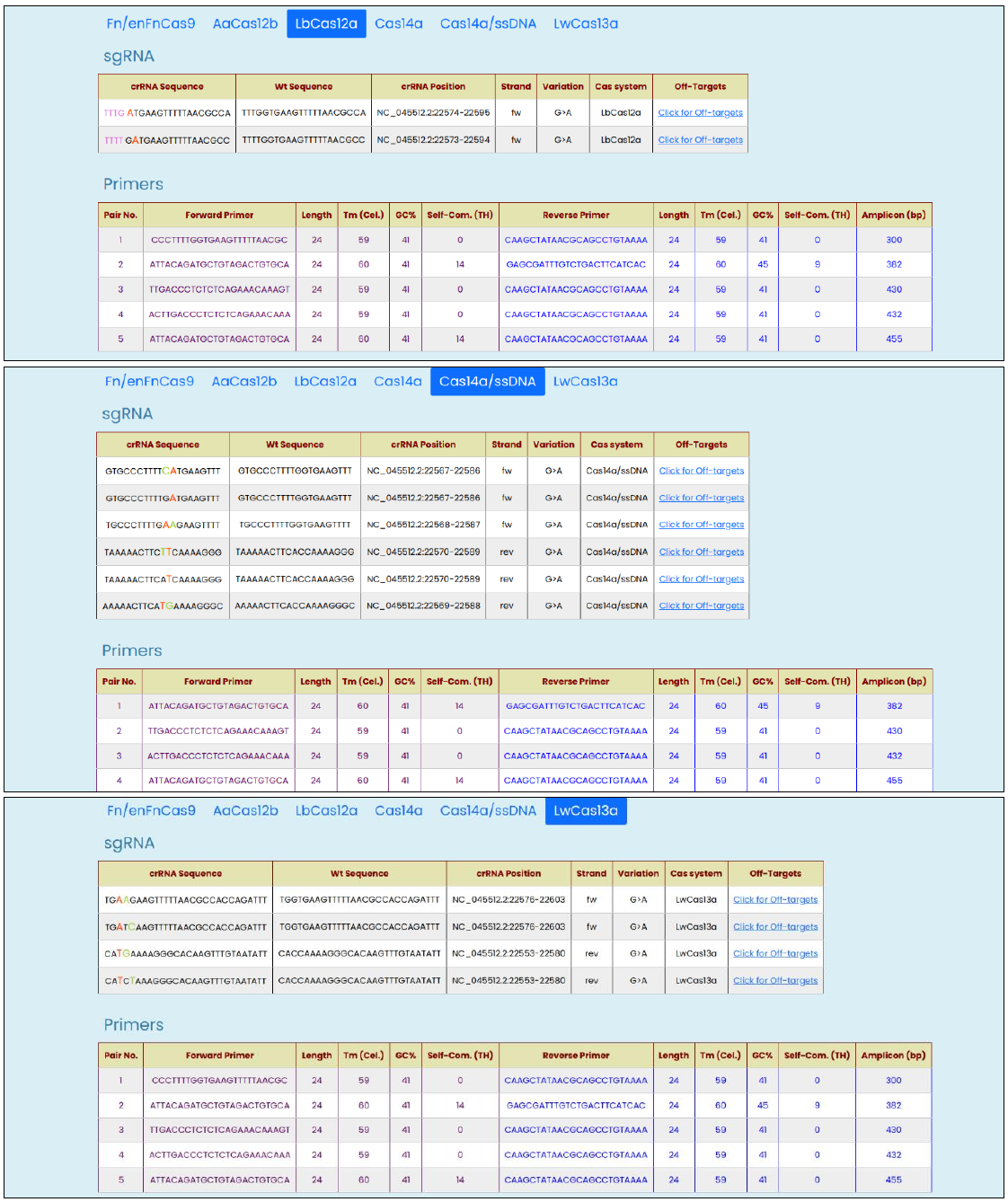
Fig. 6, Targetable SARS-CoV-2 S_G339D mutation output window with modified crRNA designed for AaCas12b and Cas14a.
iii) Seq-CriSNPr; for targeting unreported or novel human and SARS-CoV-2 SNVs
Since the previous two interfaces report curated CriSNPr crRNA information from the CriSNPr database, the outputs are immediately available but are only limited to clinically relevant human or SARS-CoV-2 SNPs/SNVs. To target any previously reported or novel SNP or SNV present in human or SARS-CoV-2 that are not previously available in CriSNPr’s database, the Seq-CriSNPr interface considers a 20 to 30 nucleotides sequence containing the SNP/SNV along with the position and nucleobase identity of SNP/SNV to be targeted within the input sequence. Using this information, Seq-CriSNPr does real-time crRNA and primer oligos design and gives out all the information required for the detection assays.
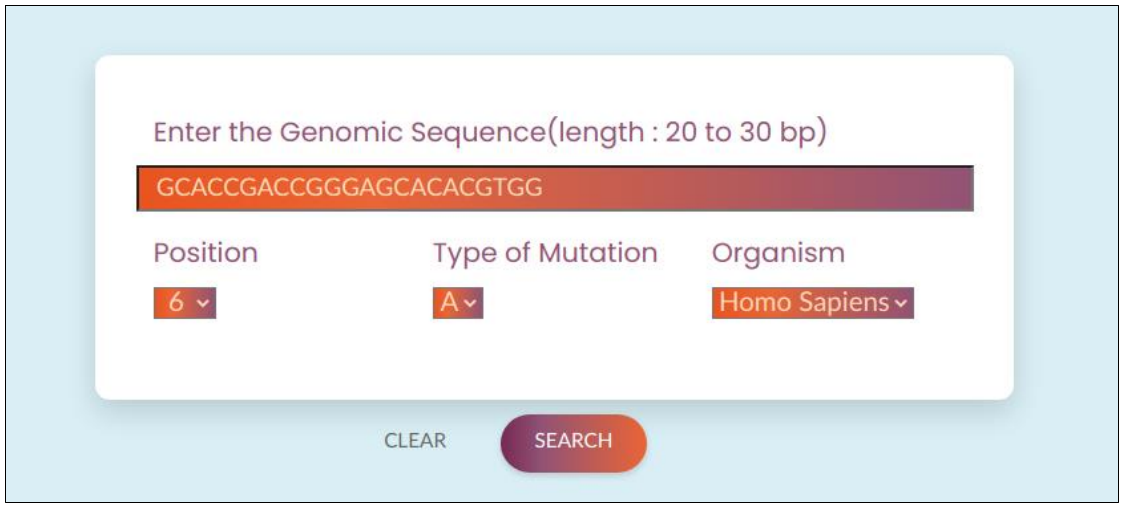
Fig. 7, Seq-CriSNPr input window with examples for user’s guide, with designated input spaces for nucleotide sequence, the position of SNV, and the nucleotide identity. The current version of Seq-CriSNPr also gives the option to choose from human and SARS-CoV-2.
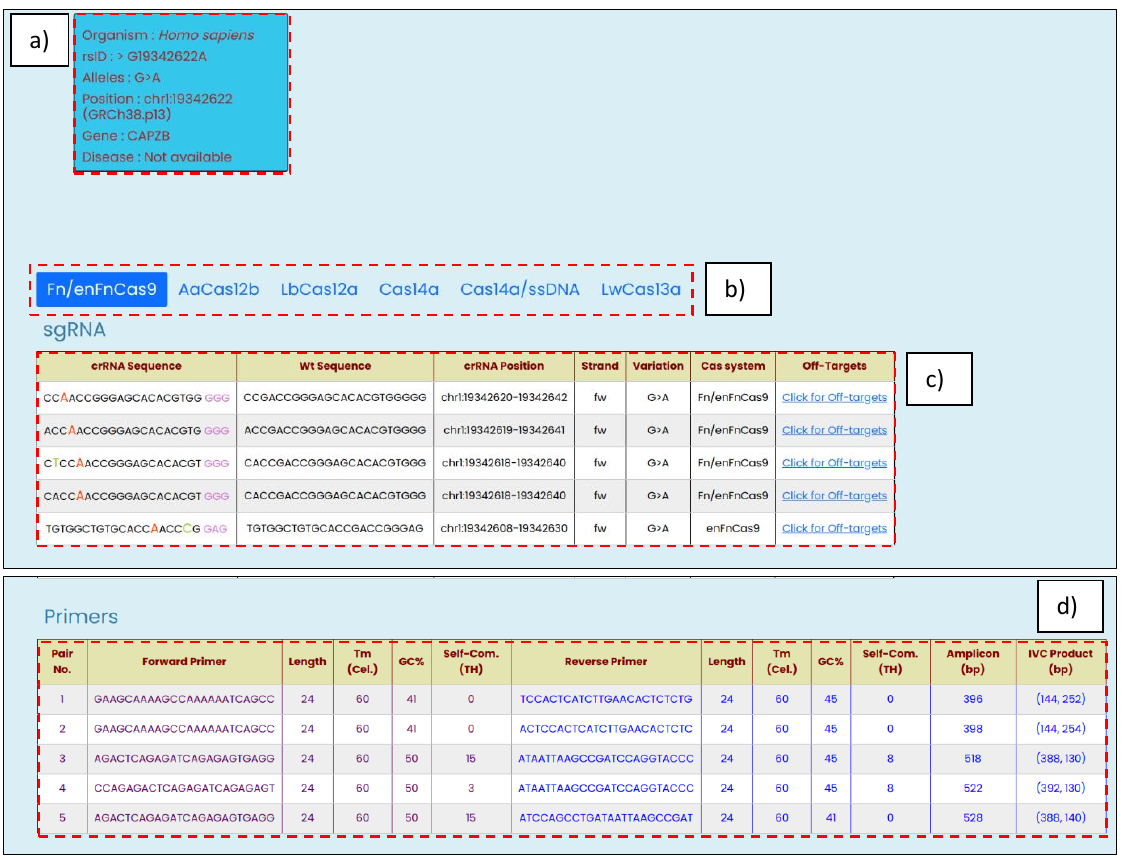
- SNV validation, with information such as an organism of origin, available dbSNP rsID, reported alleles, genome coordinates, gene ID any if any disease association.
- Cas system selection panel highlights the selected option, and results are shown for it
- SNV targeting modified crRNA sequences with other related information such as reference WT sequences, genomic positions, strand orientation, targeted allelic change, and the list of off-targets against the modified crRNA.
- Top 5 PCR amplicon primer-pair with annealing temperature, GC%, and self-complementary (TH) score as provided by the Primer3Plus
Fig. 8, Targetable non-clinically relevant human SNV output window with the following information:
Seq-CriSNPr, using the same python flask framework as of CriSNPr, gives sequence-related information similar to CriSNPr but with more user-customizable options to choose a position as well as the identity of the SNV nucleotide within a 20-30 nt query sequence.
Useful Notes
For designing IVT oligos for gRNA
As major of these gRNA designs are for in vitro single nucleobase diagnostics given below are examples for DNA oligos required for individual Cas systems utilized in previously reported protocols.
For example, target SNV rs12788013 associated with beta Thalassemia disease
a) For Fn/enFnCas9 (RAY – Rapid variant Assay, Kumar et al., 2021, Acharya et al., 2021)
Target Wt sequence: AGATATAGTGTATTTTCCCA AGG
Target Mutant sequence: AGATATAGTGTATTTTCCGA AGG
Modified crRNA sequence:AGATATAGTGTATTATCCGA
-
For SNVs to be diagnosed by the FnCas9 an additional synthetic nucleotide is added within the crRNA sequence such that it will possess two mismatches towards the target wild-type (Wt) sequences and a single mismatch towards the target mutant sequences.
The modified crRNA sequence obtained from CriSNPr can be ordered as DNA oligos with a T7-RNA Polymerase promoter sequence at the 5’ end and a part of the tracrRNA sequence at the 3’ end. If the crRNA sequence doesn’t start with a G, an additional G is required in the front to get optimal RNA conversion during in vitro transcription (IVT).
crRNA IVT forward DNA oligo:
5’ TAATACGACTCACTATAGAGATATAGTGTATTATCCGAGTTTCAGTTGCTGAATTAT 3’
17 nucleotides T7-RNA Polymerase promoter sequence
G - Additional G is only required if the crRNA doesn’t start with a G.
Yellow highlighted is the crRNA sequence designed by CriSNPr
19 nucleotides complementary tracrRNA region
And just the reverse complement sequence of the forward primer is to be ordered as a reverse primer DNA oligo.
crRNA IVT reverse DNA oligo:
5’ ATAATTCAGCAACTGAAACTCGGATAATACACTATATCTCTATAGTGAGTCGTATTA 3’
-
Equimolar annealing of forward and reverse DNA oligos is performed to assemble into a dsDNA template sequence, used for making sgRNA. The assembled template is to be used as a substrate for in vitro transcription by T7 RNA polymerase, using the T7 in vitro transcription kit by following the manufacturer’s instructions.
The CriSNPr generated modified crRNA can also be ordered as single RNA molecules with SNV targeting 20 nucleotides towards 5’ followed by a 19 nucleotide part of tracrRNA. Which, for better shelf life can be end protected by phosphorothioate bonds (*).
5’ A*G*AUAUAGUGUAUUAUCCGAGUUUCAGUUGCUGAAUU*A*U 3’Together with crRNA we will also require a tracrRNA, which can be ordered as single RNA molecules end protected by phosphorothioate bonds (*) and labeled with FAM at 3’ end.
5’ G*U*AAUUAAUGCUCUGUAAUCAUUUAAAAGUAUUUUGAACGGACCUCUGUUUGACACGUC*U*U – FAM 3’The primers for target amplicons can be ordered the same as provided by the CriSNPr
Further, a detailed protocol for the assay is available at https://elifesciences.org/articles/67130#appendix-1
b) Cas14a (DETECTR – DNA Endonuclease-Targeted CRISPR Trans Reporter, Harrington et al., 2018)
Target Wt sequence: GTGTATTTTCCCAAGGTTTG
Target Mutant sequence: GTGTATTTTCCGAAGGTTTG
Modified crRNA sequence:GTGTATTTTCCGAAGGTTTG
-
For SNVs to be diagnosed by the Cas14a the target SNV is just kept at a position of 12 and that is enough to discriminate between wild type and mutant alleles. The crRNA sequence will have one mismatch towards wild-type sequences and no mismatch towards the target mutant sequences.
Cas14a sgRNA can be ordered as RNA molecules with the following tracrRNA and combined crRNA sequences.
- To synthesize sgRNA via IVT, the modified crRNA sequence obtained from CriSNPr can be ordered as DNA oligos with a T7-RNA Polymerase promoter sequence followed by a tracrRNA sequence at the 5’ end and crRNA at the 3’ end.
5’ UUCACUGAUAAAGUGGAGAACCGCUUCACCAAAAGCUGUCCCUUAGGGGAUUAGAACUUGAG
TGAAGGUGGGCUGCUUGCAUCAGCCUAAUGUCGAGAAGUGCUUUCUUCGGAAAGUAACCCUCG
AAACAAAUUCAUUUgaaaGAAUGAAGGAAUGCAACGUGUAUUUUCCGAAGGUUUG 3’
crRNA IVT forward DNA oligo:
5’ TAATACGACTCACTATAGTTCACTGATAAAGTGGAGAACCGCTTCACCAAAAGCTGTCCCTTAGGGGATTAG
AACTTGAGTGAAGGTGGGCTGCTTGCATCAGCCTAA 3’
17 nucleotides T7-RNA Polymerase promoter sequence
G – Required additional G if not present at the beginning of the sequences to be in vitro transcribed by T7-RNA Polymerase
Cyan highlighted is the region complementary to the reverse DNA oligo, after an extension PCR together they form a single dsDNA template to be used for IVT reaction.
crRNA IVT reverse DNA oligo:
5'
GGTGGGCTGCTTGCATCAGCCTAATGTCGAGAAGTGCTTTCTTCGGAAAGTAACCCTCGAAACAAAT
TCATTTgaaaGAATGAAGGAATGCAACGTGTATTTTCCGAAGGTTTG3'
Reverse complement:
CAAACCTTCGGAAAATACACGTTGCATTCCTTCATTCtttcAAATGAATTTGTTTCGAGGGTTACTTTCCGAAGAA
AGCACTTCTCGACATTAGGCTGATGCAAGCAGCCCACC
Yellow highlighted is the crRNA sequence designed by CriSNPr
By using forward and reverse DNA oligos, an overlap extension PCR is performed to assemble smaller DNA fragments into a larger sequence, which was used for making each sgRNA.
The assembled template can be used as a substrate for in vitro transcription by T7 RNA polymerase, using the T7 in vitro transcription kit by following the manufacturer’s instructions.
Further, a detailed protocol is available at https://www.science.org/doi/abs/10.1126/science.aav4294
c) For AaCas12b (CDetection – Cas12b-mediated DNA detection, Teng et al., 2019)
Target Wt sequence:TTT TCCCAAGGTTTGAACTAGCT
Target Mutant sequence:TTT TCCGAAGGTTTGAACTAGCT
Modified crRNA sequence: TCCGAAGGTTAGAACTAGCT
-
For SNVs to be detected by the AaCas12b the target SNV is just kept at a position of 12 and that is enough to discriminate between wild type and mutant alleles. The crRNA sequence will have one mismatch towards wild-type sequences and no mismatch towards the target mutant sequences.
AaCas12b sgRNA can be ordered as RNA molecules with the following tracrRNA and combined crRNA sequences.
- To synthesize sgRNA via IVT, the modified crRNA sequence obtained from CriSNPr can be ordered as DNA oligos with a T7-RNA Polymerase promoter sequence followed by a tracrRNA sequence at the 5’ end and crRNA at the 3’ end.
5’ 5’GUCUAAAGGACAGAAUUUUUCAACGGGUGUGCCAAUGGCCACUUUCCAGGUGGCAAAGCCC
GUUGAACUUCAAGCGAAGUGGCAC
UCCGAAGGUUAGAACUAGCU 3’
crRNA IVT forward DNA oligo:
5’ TAATACGACTCACTATAGTCTAAAGGACAGAATTTTTCAACGGGTGTGCCAATGGCCACTTTCCAGGTGGCAAAGCCCGTTG 3’
17 nucleotides T7-RNA Polymerase promoter sequence
Cyan highlighted is the region complementary to the reverse DNA oligo, after an extension PCR together they form a single dsDNA template to be used for IVT reaction.
crRNA IVT reverse DNA oligo:
CCAGGTGGCAAAGCCCGTTGAACTTCAAGCGAAGTGGCACTCCGAAGGTTAGAACTAGCT
Reverse complement:
AGCTAGTTCTAACCTTCGGAGTGCCACTTCGCTTGAAGTTCAACGGGCTTTGCCACCTGG
Yellow highlighted is the crRNA sequence designed by CriSNPr.
By using forward and reverse DNA oligos, an overlap extension PCR is performed to assemble smaller DNA fragments into a larger sequence, which was used for making each sgRNA.
The assembled template can be used as a substrate for in vitro transcription by T7 RNA polymerase, using the T7 in vitro transcription kit by following the manufacturer’s instructions.
Further, a detailed protocol is available at https://link.springer.com/article/10.1186/s13059-019-1742-z
The amplified WT and mutant nucleotide sequences when treated with modified gRNA:Cas complexes can be distinguished by adapting to different readouts such as gel electrophoresis, capillary electrophoresis, fluorescence measurement, or lateral flow assay, etc (1-7 ).
Frequently Asked Questions
I am unable to design crRNAs for my variant id (rsID or mutation id in the case of SARS-CoV-2)
If the server is not able to design crRNA for any variation, one can try using a sequence-based query through Seq-CriSNPr. Even though the CriSNPr database is updated on a regular basis, it might miss variant ids that are recently updated in ClinVar or the dbSNP database. Please mail us to include your variations in the database.
I want to design crRNA for organisms other than SARS-CoV-2 or humans
One can mail us to include your genome of interest. We will try to incorporate the variation database of organisms into the CriSNPr, if available. Otherwise, the genome can be included in Seq-CriSNPr and can design crRNAs and primers through a sequence-based query.
Why are there two tabs of Cas14 in the result section?
Cas14 requires TTTA PAM when cleaving a dsDNA target sequence, but is not dependent on any PAM when it comes to an ssDNA target sequence. Based on the availability and uses end, both are included in the result section.
On what basis the crRNA provided are ranked?
Currently, there is no scoring algorithm developed to score these SNV targeting modified crRNA because of limited experimentally validated data. But one can still consider crRNA sequences with no or the minimum number of off-targets.
On what basis are the provided amplification primers ranked?
CriSNPr uses Primer3 python library for designing the provided primer sequences and is ranked by considering different parameters such as annealing temperature, GC%, and self-complementary (TH) score as provided by the Primer3Plus. To reduce the non-specific amplification by primer sequences, these were further filtered for off-targets on a representative bacterial genome database (NCBI), virus genome database (NCBI), and human genome/transcriptome (GENCODE GRCh38) with up to 2 mismatches.
Regarding any error or trouble in designing crRNA/primers targeting a particular SNV, please raise the issue to the https://github.com/asgarhussain/CriSNPr or direct mail to asgar.hussain@igib.in.
CriSNPr crRNA database link : Human/Sars-Cov-2 variants database
References:
Myhrvold, Cameron, et al. "Field-deployable viral diagnostics using CRISPR-Cas13." Science 360.6387 (2018): 444-448.
Li, Shi-Yuan, et al. "CRISPR-Cas12a-assisted nucleic acid detection." Cell discovery 4.1 (2018): 1-4.
-
Harrington, Lucas B., et al. "Programmed DNA destruction by miniature CRISPR-Cas14 enzymes." Science 362.6416 (2018): 839-842.
-
Teng, Fei, et al. "CDetection: CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity." Genome biology 20.1 (2019): 1-7.
-
Azhar, Mohd, et al. "Rapid and accurate nucleobase detection using FnCas9 and its application in COVID-19 diagnosis." Biosensors and Bioelectronics 183 (2021): 113207.
-
Kumar, Manoj, et al. "FnCas9-based CRISPR diagnostic for rapid and accurate detection of major SARS-CoV-2 variants on a paper strip." Elife 10 (2021): e67130.
-
Acharya, Sundaram, et al. "Engineered PAM-flexible FnCas9 variants for robust and specific genome editing and diagnostics." (https://doi.org/10.21203/rs.3.rs-990232/v1, 2021).
-
Untergasser, Andreas, et al. "Primer3—new capabilities and interfaces." Nucleic acids research 40.15 (2012): e115-e115.
-
Bae, Sangsu, Jeongbin Park, and Jin-Soo Kim. "Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases." Bioinformatics 30.10 (2014): 1473-1475.